Priors for Bayesian analysis
Astromodels supports the definition of priors for all parameters in your model. You can use as prior any function (although of course not all functions should be used this way, but the choice is up to you).
First let’s define a simple model containing one point source (see the “Model tutorial” for more info):
[1]:
%%capture
from astromodels import *
# Create a point source named "pts1"
pts1 = PointSource('pts1',ra=125.23, dec=17.98, spectral_shape=Powerlaw())
# Create the model
my_model = Model(pts1)
Now let’s assign uniform priors to the parameters of the powerlaw function. The function uniform_prior is defined like this:
[2]:
Uniform_prior.info()
- description: A function which is constant on the interval lower_bound - upper_bound and 0 outside the interval. The extremes of the interval are counted as part of the interval.
- formula: $ f(x)=\begin{cases}0 & x < \text{lower_bound} \\\text{value} & \text{lower_bound} \le x \le \text{upper_bound} \\ 0 & x > \text{upper_bound} \end{cases}$
- default parameters:
- lower_bound:
- value: 0.0
- desc: Lower bound for the interval
- min_value: -inf
- max_value: inf
- unit:
- is_normalization: False
- delta: 0.1
- free: True
- upper_bound:
- value: 1.0
- desc: Upper bound for the interval
- min_value: -inf
- max_value: inf
- unit:
- is_normalization: False
- delta: 0.1
- free: True
- value:
- value: 1.0
- desc: Value in the interval
- min_value: None
- max_value: None
- unit:
- is_normalization: False
- delta: 0.1
- free: True
- lower_bound:
We can use it as such:
[3]:
# Set 'lower_bound' to 0, 'upper bound' to 10, and leave the 'value' parameter
# to the default value
pts1.spectrum.main.Powerlaw.K.prior = Uniform_prior(lower_bound = 0, upper_bound=10)
# Display it
pts1.spectrum.main.Powerlaw.K.display()
spectrum
main
Powerlaw
K
lower_bound
upper_bound
value
spectrum
main
Powerlaw
K
_ipython_canary_method_should_not_exist_
_ipython_display_
_ipython_canary_method_should_not_exist_
_repr_mimebundle_
_ipython_canary_method_should_not_exist_
_ipython_canary_method_should_not_exist_
_repr_markdown_
_ipython_canary_method_should_not_exist_
_repr_svg_
_ipython_canary_method_should_not_exist_
_repr_png_
_ipython_canary_method_should_not_exist_
_repr_pdf_
_ipython_canary_method_should_not_exist_
_repr_jpeg_
_ipython_canary_method_should_not_exist_
_repr_latex_
_ipython_canary_method_should_not_exist_
_repr_json_
_ipython_canary_method_should_not_exist_
_repr_javascript_
Parameter K = 1.0 [1 / (cm2 keV s)]
(min_value = 1e-30, max_value = 1000.0, delta = 0.1, free = True) [prior: Uniform_prior]
Now, lets’s set a Gaussian prior on the spectral index
[4]:
pts1.spectrum.main.Powerlaw.index.prior = Gaussian(mu=-2, sigma=1)
pts1.spectrum.main.Powerlaw.index.display()
spectrum
main
Powerlaw
index
F
mu
sigma
spectrum
main
Powerlaw
index
_ipython_canary_method_should_not_exist_
_ipython_display_
_ipython_canary_method_should_not_exist_
_repr_mimebundle_
_ipython_canary_method_should_not_exist_
_ipython_canary_method_should_not_exist_
_repr_markdown_
_ipython_canary_method_should_not_exist_
_repr_svg_
_ipython_canary_method_should_not_exist_
_repr_png_
_ipython_canary_method_should_not_exist_
_repr_pdf_
_ipython_canary_method_should_not_exist_
_repr_jpeg_
_ipython_canary_method_should_not_exist_
_repr_latex_
_ipython_canary_method_should_not_exist_
_repr_json_
_ipython_canary_method_should_not_exist_
_repr_javascript_
Parameter index = -2.01 []
(min_value = -10.0, max_value = 10.0, delta = 0.20099999999999998, free = True) [prior: Gaussian]
[5]:
# Let's get 500 points uniformly distributed between -20 and 20
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
from jupyterthemes import jtplot
jtplot.style(context="talk", fscale=1, ticks=True, grid=False)
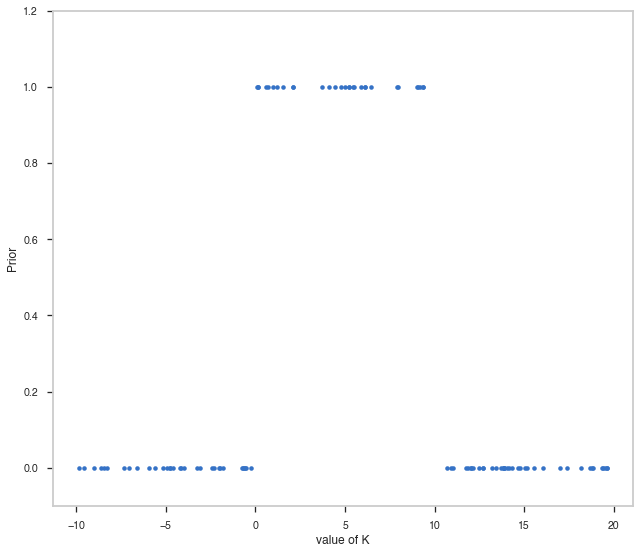
random_points = np.random.uniform(-10,20,100)
fig, ax = plt.subplots()
ax.plot(random_points,pts1.spectrum.main.Powerlaw.K.prior(random_points), '.' )
ax.set_ylim([-0.1,1.2])
ax.set_xlabel("value of K")
ax.set_ylabel("Prior")
spectrum
main
Powerlaw
K
[5]:
Text(0, 0.5, 'Prior')
[6]:
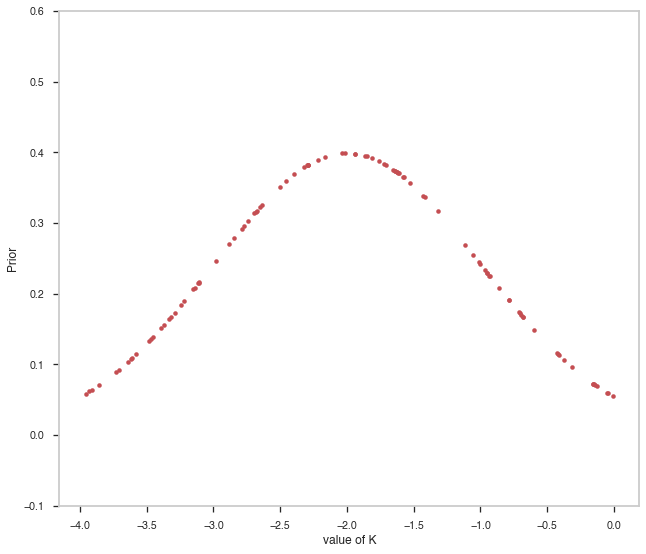
random_points = np.random.uniform(-4,0,100)
fig, ax = plt.subplots()
ax.plot(random_points,pts1.spectrum.main.Powerlaw.index.prior(random_points), 'r.' )
ax.set_ylim([-0.1,0.6])
ax.set_xlabel("value of K")
ax.set_ylabel("Prior")
spectrum
main
Powerlaw
index
[6]:
Text(0, 0.5, 'Prior')
[ ]: